DNA Replication
This article covers:
What is DNA replication?
Why is it important?
When does it take place?
Where does it take place?
Steps of the DNA replication and the enzymes involved.
Basic information
DNA replication is a process where the cell makes exact copies of the DNA.
Why is it important?
Cell division is a very important process in a living organism. It helps in the growth and development of an organism. DNA is a vital biomolecule that carries all the information needed for a fully functional cell. Cell division leads to the formation of two daughter cells. DNA replication makes sure that the daughter cells have the same amount of DNA and that all the genetic information is copied.
Where and when does it take place?
In eukaryotic cells, DNA replication takes place in the Nucleus of the cell in the S phase of the cell cycle. This is before the cell division process.
Steps involved in DNA replication
DNA replication always takes place in the 5' - 3' direction. This means that the new nucleotides are added at the 3' end of the free end (Figure 1) and therefore the synthesis of the strand goes from the 5' - 3' end.
DNA replication consists of three major steps. Namely
Initiation
Elongation
Termination
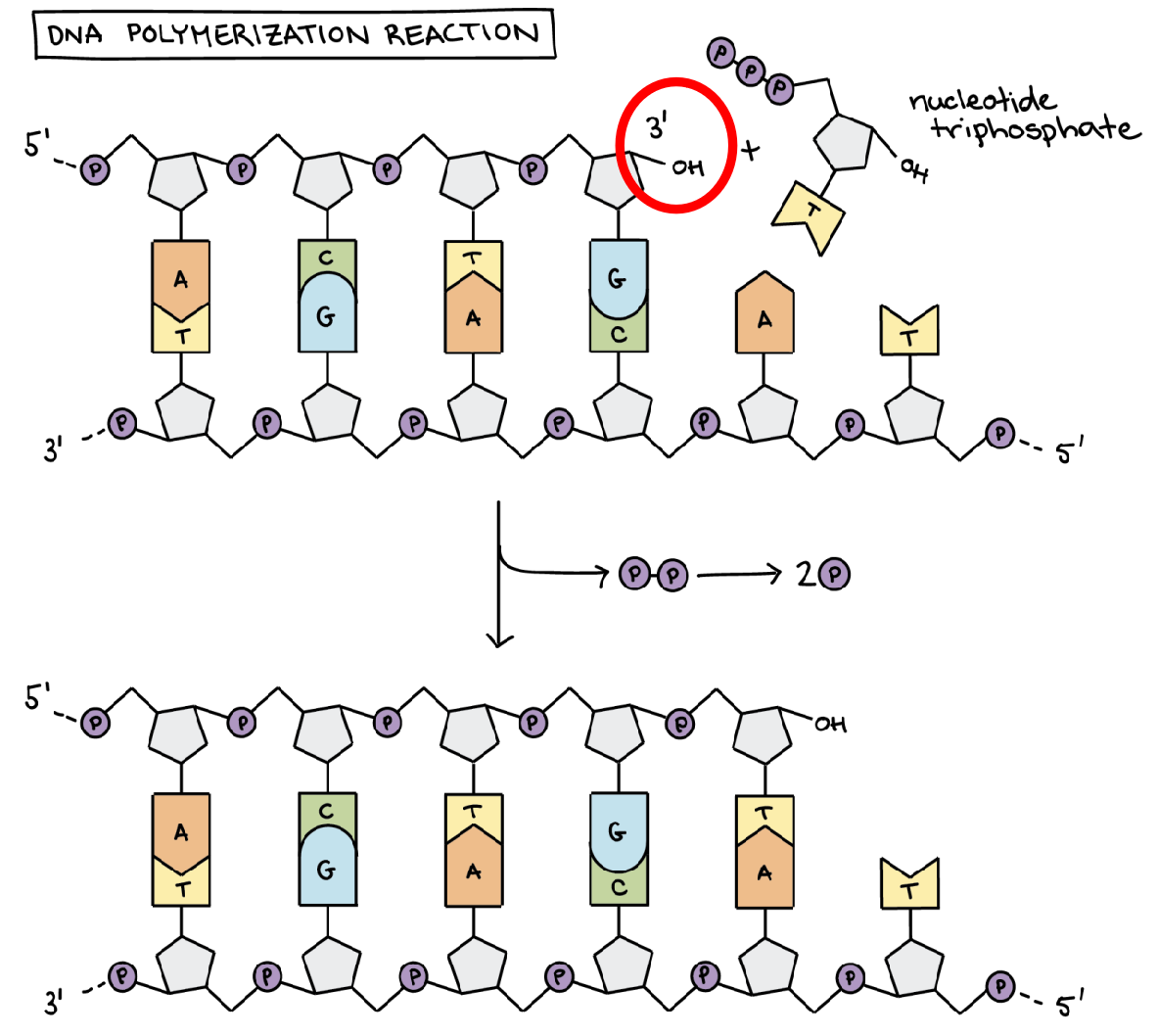
Figure 1
Steps of the DNA replication:
Step 1: Initiation
This process takes place at the 'Origin of replication' regions. These regions are usually rich in Adenine and Thymine (A-T regions) because it is easier to break the double hydrogen bond between A-T compared to the triple hydrogen bond between Guanine and Cytosine (G-C). There can be multiple Origin of replication (ORI) sites in the DNA.
Initiation is started by the Pre-replication protein complex (Pre-RC). It binds at the ORI and breaks the hydrogen bonds between a few bases. This protein complex controls the starting of the replication process (Figure 2).
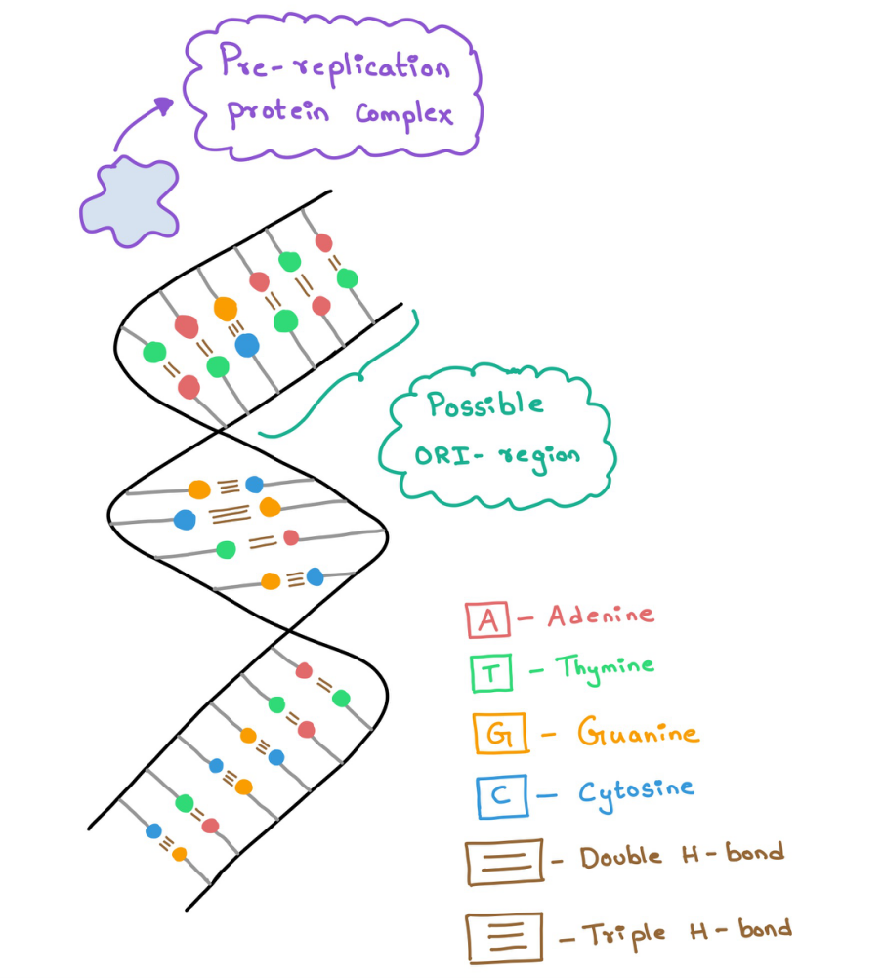
Figure 2
DNA Helicase is the enzyme that unwinds the DNA leading to a replication bubble. It breaks the hydrogen bond between the bases and separates the nucleotides. It requires a lot of ATP (Adenosine triphosphate) as the energy source (Figure 3).
Once the nucleotides are separated to form two single strands of DNA, there are chances that they might bind again and form a double strand. This is called Re-annealing. They can also be damaged by the Nucleases in the cell. The above two processes are prevented by the Single-strand binding proteins (SSBP) they bind to the single-stranded DNA and prevent them from re-annealing and protect them from the nucleases (Figure 3).
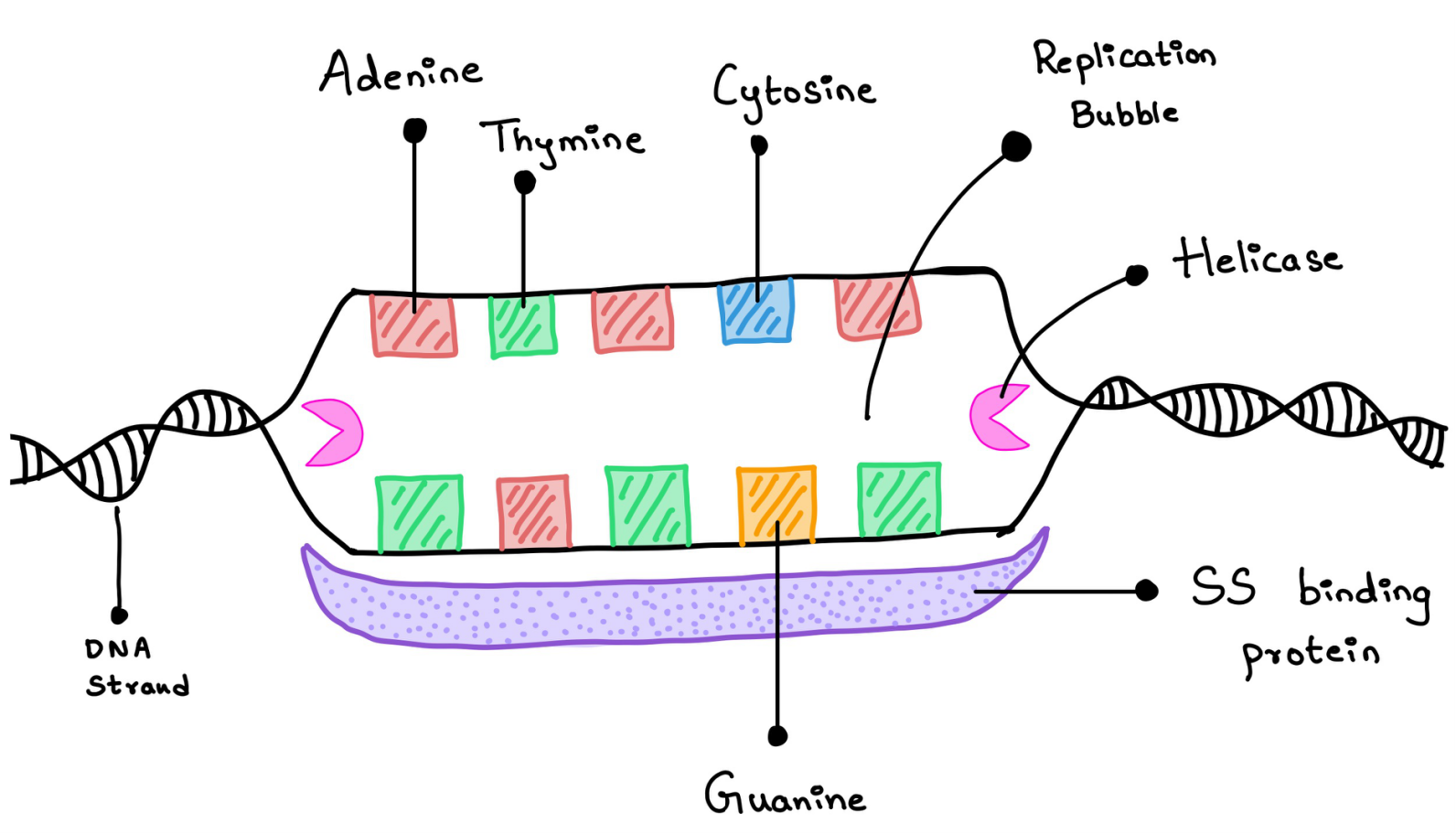
Figure 3
As the helicase separates the DNA strands and pushes through them, the DNA strand ahead of it tends to bunch up and supercoil. This causes hindrance to helicase movement and activity (Figure 4).
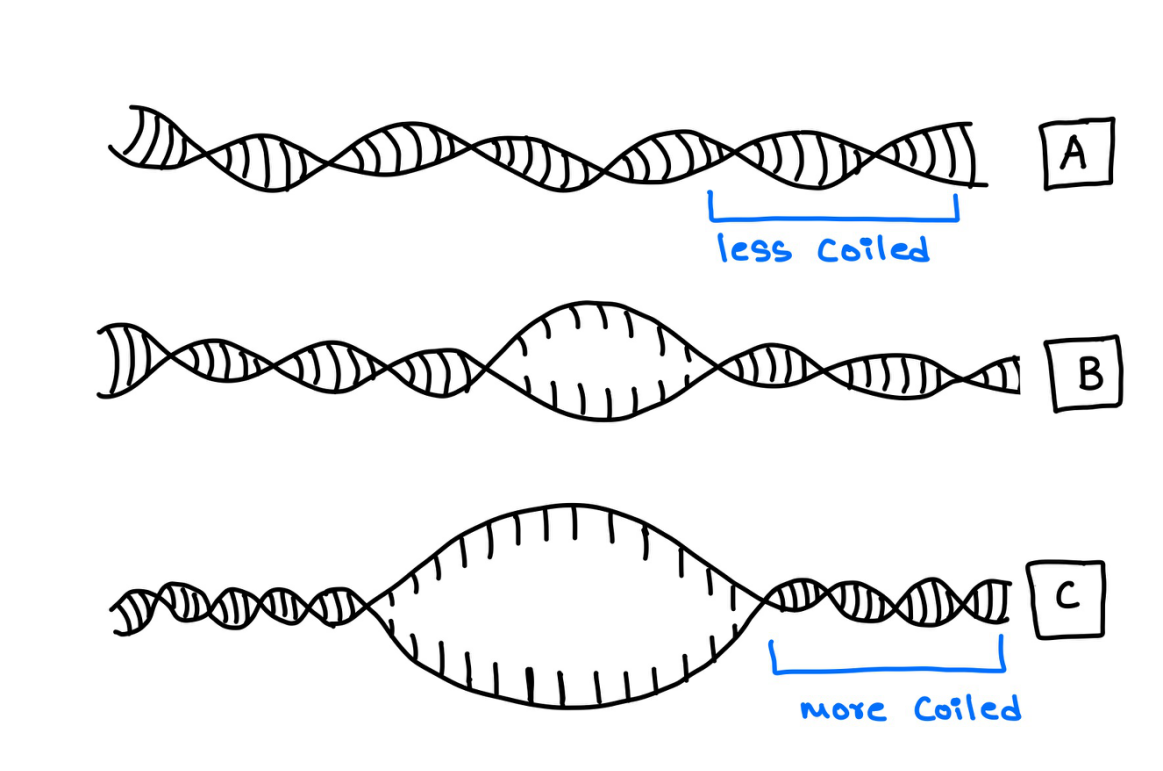
Figure 4
Topoisomerases are the enzymes that reduce this hindrance (Figure 5). They do so by cutting a part of the DNA strand to reduce the coiling and re-sealing that part of the DNA. Topoisomerases use the nuclease arm to cut the DNA and the ligase arm to seal them back (Figure 6). Even DNA Gyrase is involved in relaxing the DNA supercoil.
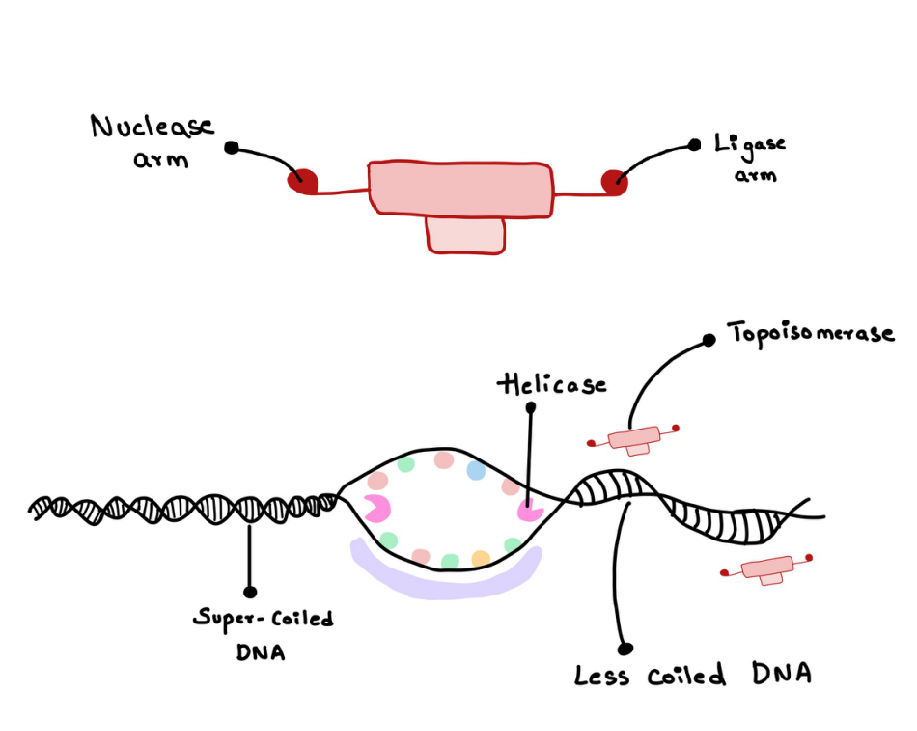
Figure 5
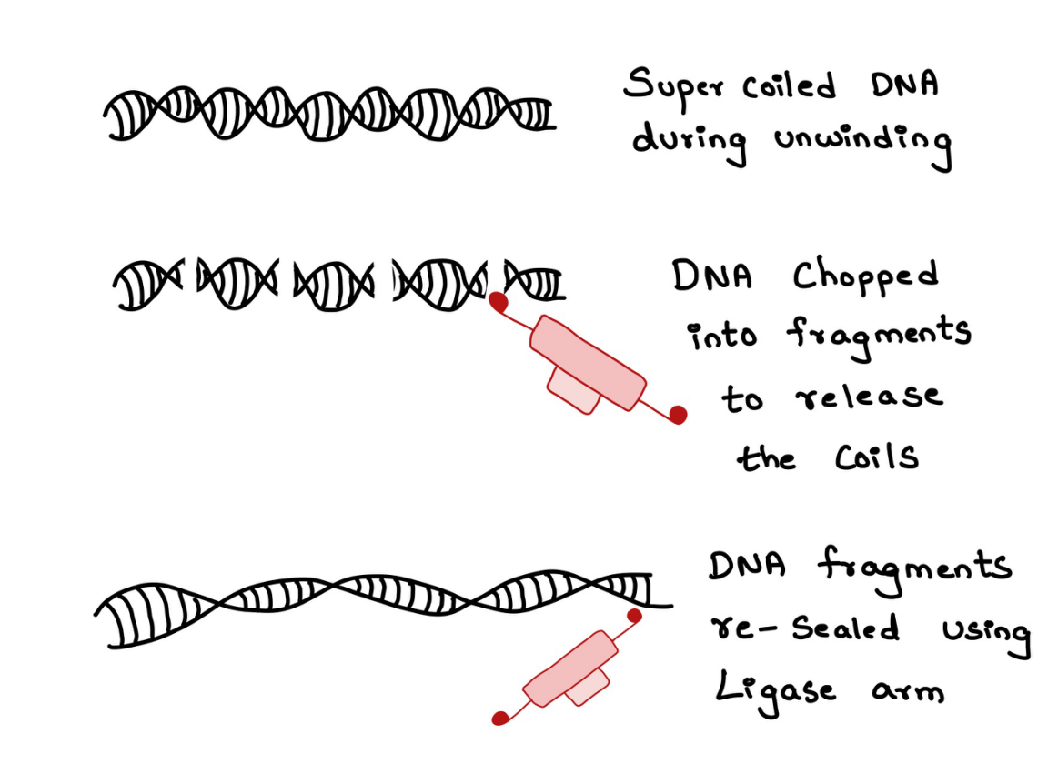
Figure 6
Nucleases - Destructive enzymes that eat up the DNA strand by breaking phosphodiester bonds
After the initiation process, the DNA strands are separated to form two single strands, and a replication bubble is formed.
Step 2: Elongation
This process involves synthesizing new strands of DNA using the parental strands (Figure 7). These parental strands are separated and complementary strands are synthesized keeping the two parental strands as a reference. But, the process is not the same for both the strands.
Concept of leading and lagging strand :
One of the strands of the parental DNA is 5' to 3' and the other strand is 3' to 5' (Figure 7). Let us call them the 5' to 3' parental strand and the 3' to 5' parental strand respectively. The new strands are synthesized accordingly. For the 5' to 3' parental strand, the complementary strand should be 3' to 5' and for the 3' to 5' parental strand the complimentary strand should be 5' to 3'.
It is known that the synthesis of DNA strands takes place only in the 5' to 3' direction because new nucleotides can be added in the 3' end (refer above part). Therefore, the complementary to the parental strand running 3' to 5' can be easily synthesized and this is called the Leading strand. But, this is not the case for the parental strand 5' to 3' and it is called the Lagging strand (Figure 8).
There are three enzymes involved in this step and they are DNA polymerase 1, DNA polymerase 3, and RNA primase.
Replication in the leading strand - RNA primase adds a short strand of nucleotides in the leading strand (Figure 9). This is to initiate the elongation process. The primase ends with a 3' end which is later continued by the DNA polymerase 3 and it keeps adding the nucleotides (Figure 10). DNA polymerase 3 also does proofreading.
Primers - they are RNA fragments and not DNA.
Proofreading - The process of recognizing wrongly paired nucleotides and replacing them with the right ones. It is done using exonuclease activity. The wrong nucleotides are replaced with the right ones.
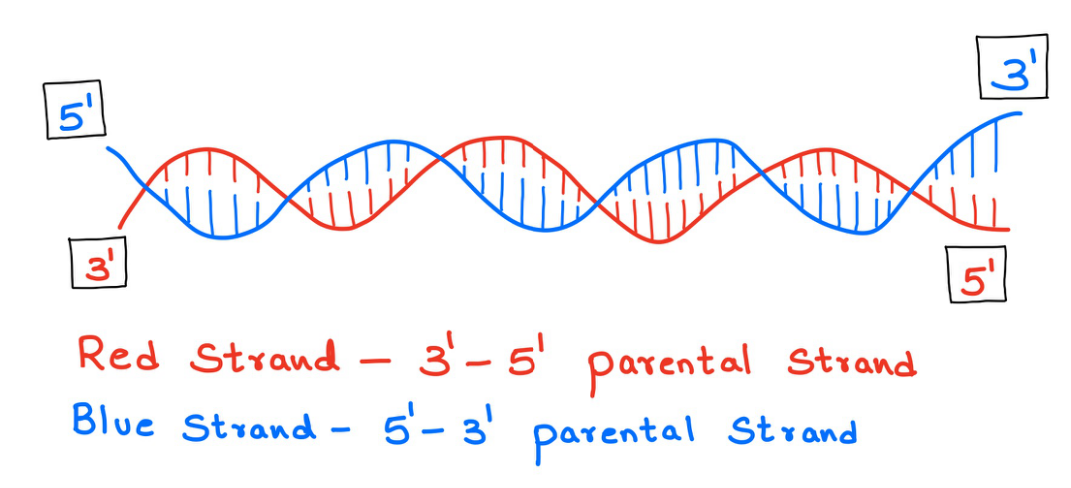
Figure 7
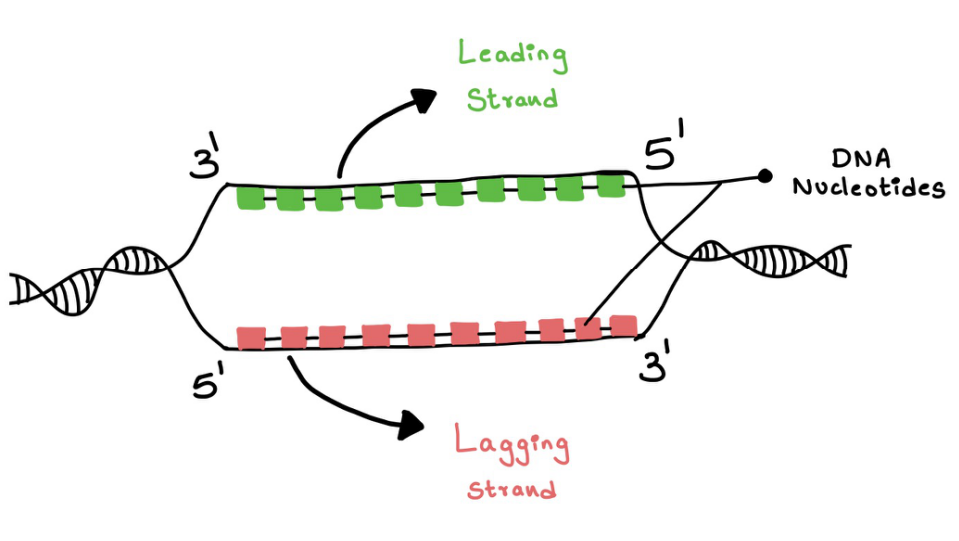
Figure 8
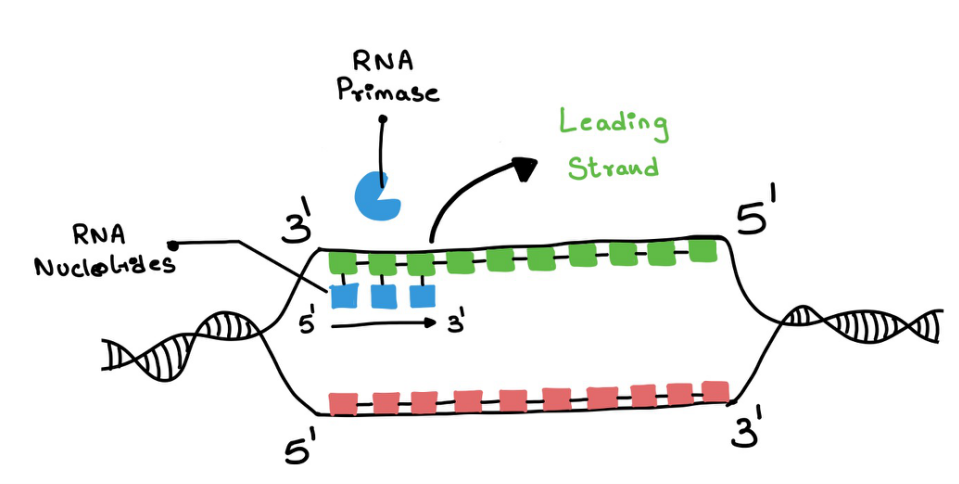
Figure 9
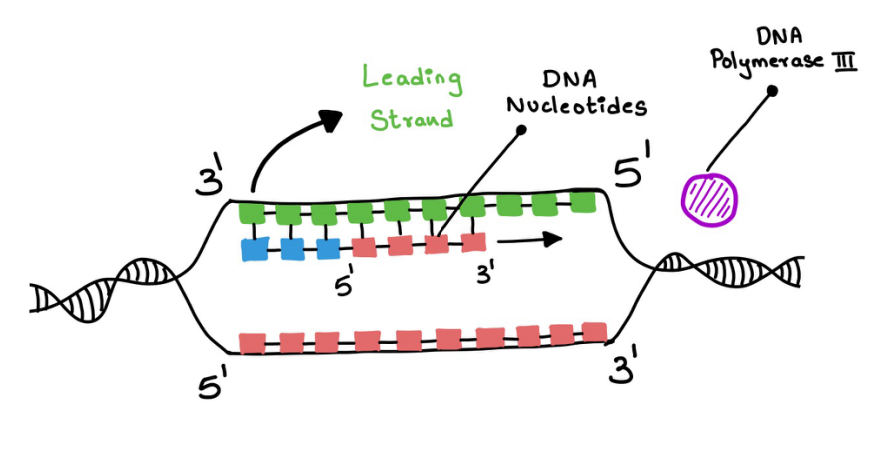
Figure 10
Replication of the lagging strand - RNA primase again adds a short strand like it did before. And DNA polymerase 3 keeps adding the nucleotides but the helicase keeps unwinding the DNA and now the lagging strand will need more primers to replicate the remaining part that is freshly unwinded again. This leads to the fragments DNA and RNA and this is called Okazaki fragments (Figure 11).
DNA polymerase 1 enzyme cuts off the RNA primers and replaces them with the DNA fragments to make it a uniform DNA strand.
It removes the RNA strands with the help of Exonuclease activity.
After the DNA pol 1 plucks off RNA primers and replaces them with DNA, and leads to gaps in the strands (Figure 12). This is fixed with the help of the DNA ligase enzyme. It joins the DNA strands together. (figure 13)
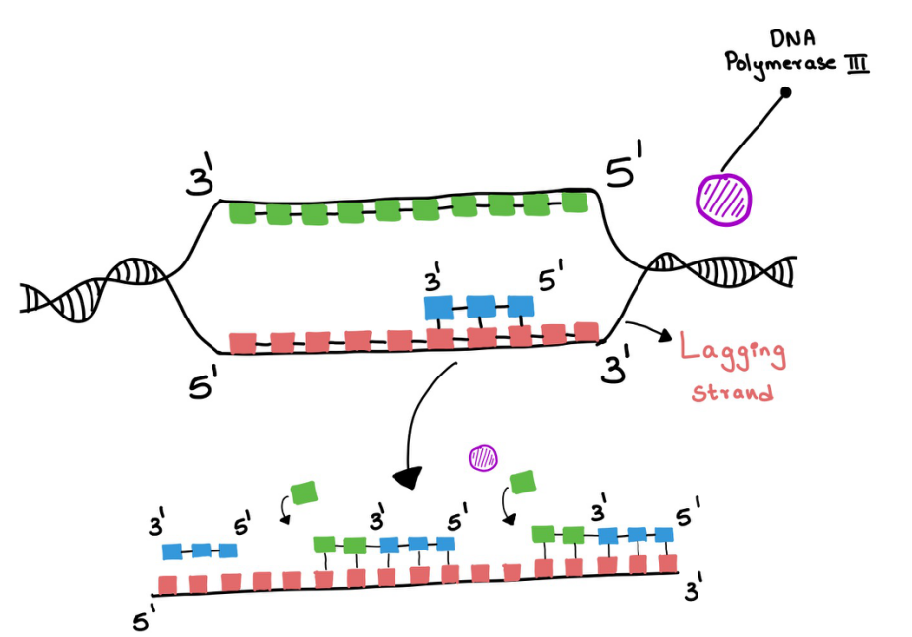
Figure 11
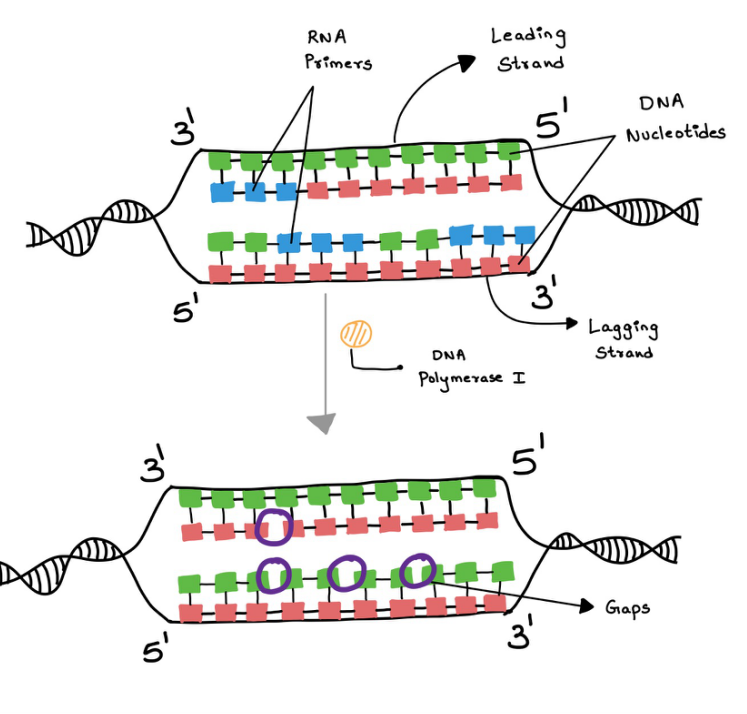
Figure 12
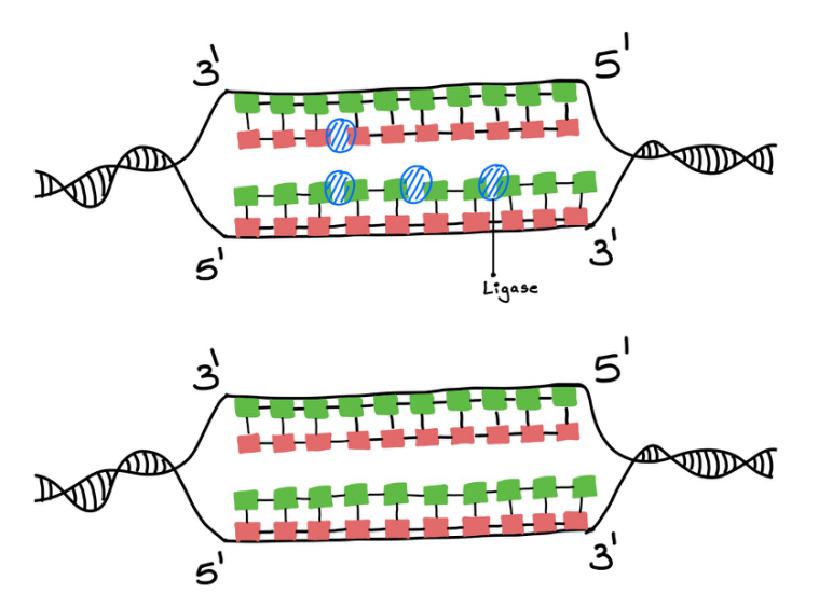
Figure 13
Step 3: Termination
Replication of DNA starts at many ORI places. This involves a lot of helicases and all the other enzymes working on the same strand of the DNA.
Termination of replication happens when two replication forks meet each other and DNA helicases bump into each other (Figure 14).
After the termination, DNA replication is complete and cell division can take place.
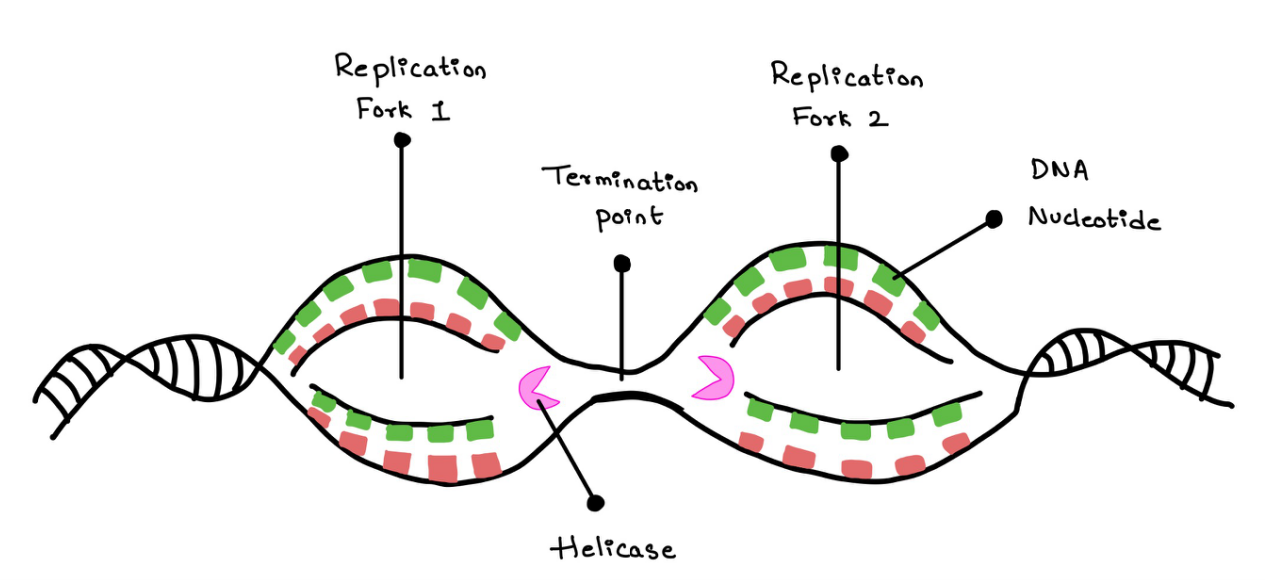
Figure 14
Sl. No | Enzymes/proteins involved | Function |
|---|---|---|
1 | Pre replication protein complex (Pre-RC) | Binds at the ORI and breaks the hydrogen bonds between a few bases |
2 | DNA helicase | Unwinds the DNA leading to a replication bubble |
3 | Single strand binding proteins (SSBP) | Bind to the single stranded DNA and prevent them from re-annealing and protect them form the nucleases |
4 | Topoisomerases, DNA Gyrase | Relax DNA supercoiling |
5 | RNA Primase | Adds a short strand of RNA nucleotides |
6 | DNA Polymerase 3 | Adds DNA nucleotides to the strand |
7 | DNA Polymerase 1 | Cuts off the RNA primers and replaces them with the DNA fragments |
8 | DNA Ligase | Joins the DNA strands |